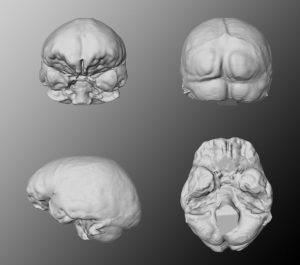
Virtual endocasts can be generated similar to the filling of cavities as we know it from natural endocasts of the braincase. A virtual endocast can be used to study the shape, the surface morphology (convolutions, imprints of blood vessels, etc.) or to calculate the endocranial capacity.
Generally, an endocast is a cast of the endocranium, the interior of the braincase and not a cast of the brain itself. The endocranium contains other components than the brain too, such as meninges, cerebrospinal fluid, blood vessels, venous sinuses and cranial nerves.
Segmentation
Producing a virtual endocast includes the segmentation of the stack of parallel cross-sectional images (slices) obtained by CT scanning. Segmentation as described above, is a mapping of symbolic information from voxel space into an anatomical name space. A predefined value is assigned to all voxels inside of the endocranium on every slice of the CT volume file, either manual or (semi-)automated. Manual segmentation needs a-priori anatomical knowledge to decide where the boundary between bone and endocranial cavity should be drawn. Automated image segmentation by grayscale or color thresholding considers voxels within a defined grayscale range as part of the virtual endocast. The exposed endocast can serve to determine the endocranial capacity and provide data on size, shape, asymmetry, patterns of arterial and venous vessels, cranial venous sinuses, and the pattern of convolutions (gyri and sulci). Additionally, the geometry can be analyzed using three-dimensional coordinates from internal landmarks.
Applications of Virtual endocasts
Virtual endocasts are especially useful if the endocranial cavity is inaccessible (complete cranium or filled with matrix) or if manual measurements are difficult to apply as in the case of fragmented fossils. For instance, the male A. africanus cranium Stw 505 was believed to exceed 600 ccm cranial capacity which would have been exceptionally high for an Australopithecine. Due to its distortions and incompleteness, traditional measurements were prone to high inter-observer differences. In 1998, Conroy et al. addressed the problem by producing virtual endocasts of the hemicranium that allowed much better control of uncertainties. Their results independently achieved in two different labs yielded a volume of 515 ccm which is still the highest for this taxon but not surprisingly high for a large male specimen. As this example shows, reproducibility and control of manipulation are main advantages working with virtual objects. Recent works additionally use reference-based reproducible methods for reconstruction of missing parts and correction of deformation.
Electronic preperation
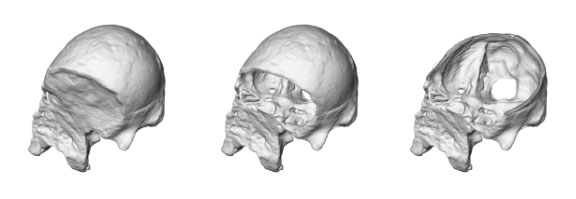
Before a cranium can be examined, electronic preparation is eventually needed. Electronic preparation is based upon segmentation as well. Slice by slice, the boundary must be set between fossilized bone and encrustation matrix. As in all VA techniques, this does not risk the precious original specimen. The task is difficult, and often requires several stages of image filters to achieve appropriate results. For example, Prossinger et al. have effectively applied a series of filter algorithms to the CT scans of the Steinheim cranium (Homo heidelbergensis) to remove the encrustations. The extent of deformation could then be assessed before endocranial volume determination, and the otherwise inaccessible frontal and sphenoid sinuses could be analyzed and compared with other specimens.